"Divide each difficulty into as many parts as is feasible and necessary to resolve it."
-René Descartes
A systems approach for studying hematopoiesis
The human blood system and its influence on health and disease is highly complex. Our blood consists of a diverse array of cell types that arise from a hematopoietic stem cell (HSC) through a tightly regulated process known as hematopoiesis. Proper functioning of the blood system is essential for maintaining overall health, as blood cells play critical roles in oxygen transport, immune defense, and tissue repair. Dysregulation of hematopoiesis can lead to a variety of blood disorders, including anemia, immunodeficiencies, and hematologic malignancies. Understanding the intricate processes governing hematopoiesis is crucial for developing effective therapies for these conditions, especially in the aging population.





Research Projects
Introduction
Ediriwickrema et al. Blood 2023
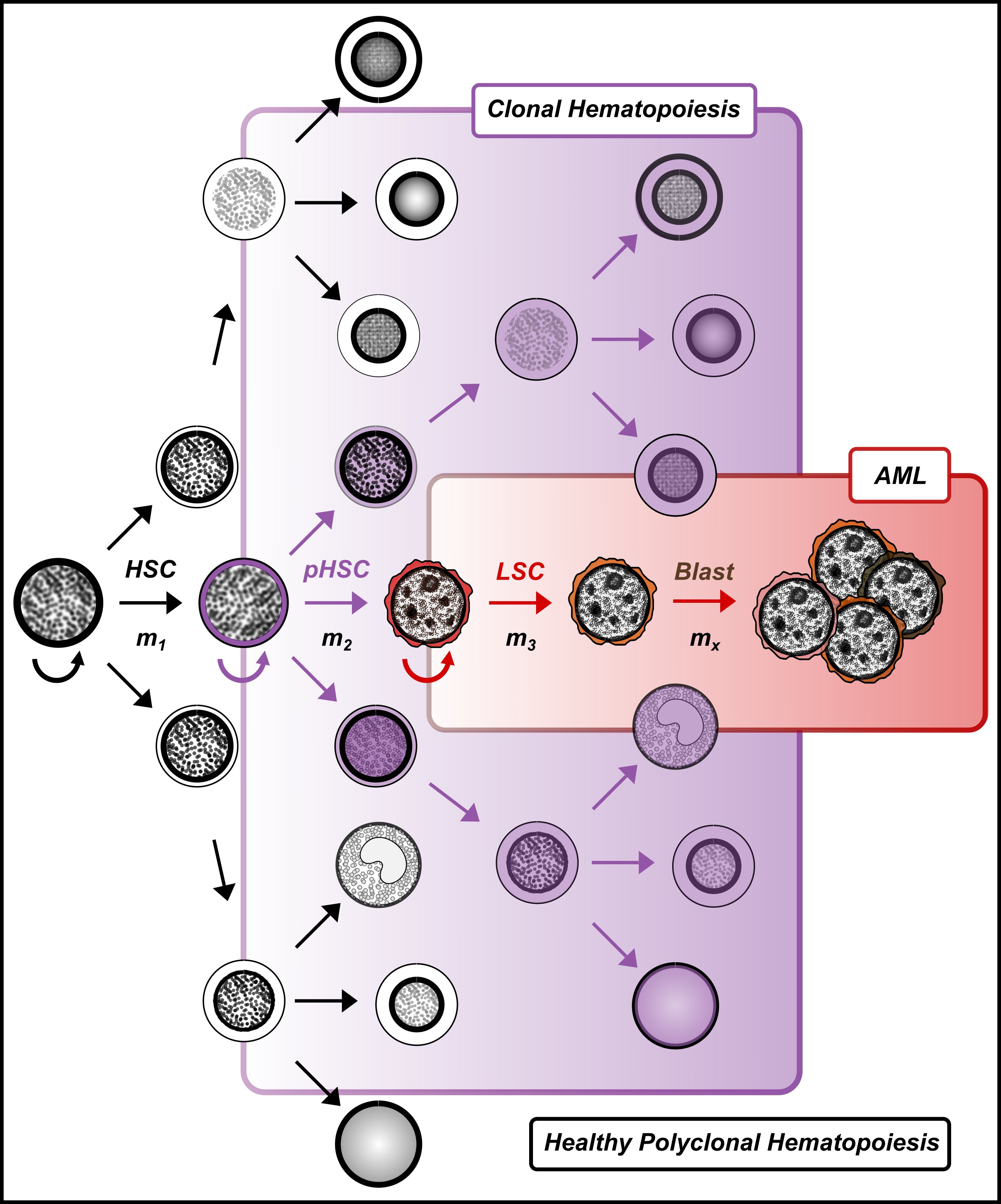
Myeloid neoplasms are heterogenous, clonal disorders arising from the malignant transformation of healthy hematopoietic stem and progenitor cells (HSPCs). The heterogeneity is due to the underlying clonal diversity of the disease and the variable functional properties of these clones. Therefore, it is not surprising that myeloid neoplasms have been historically described as syndromes with mostly ineffective treatment strategies. The inability to completely characterize the cellular architecture of myeloid neoplasms and link it to outcomes has limited our ability to manage and cure patients. Recent advancements in single cell technologies [Ediriwickrema et al. Blood 2023] (Figure 1) can provide methods for addressing these limitations by allowing researchers to characterize blood cells with high resolution. However, as the field of single cell genomics expands at unprecedented rates, it is increasingly important to develop a rigorous framework for interpreting and validating important single cell observations with orthogonal computational and functional studies to provide meaningful services for patients. We have developed such a framework for studying normal and malignant hematopoiesis which has allowed us to identify new HSPCs in adult hematopoiesis [Ediriwickrema et al. Cell Reports 2025] (Figure 2), and clinically high-risk leukemia stem cells (LSCs) in adult acute myeloid leukemia (AML) at high resolution.

Ediriwickrema et al. Cell Reports 2025
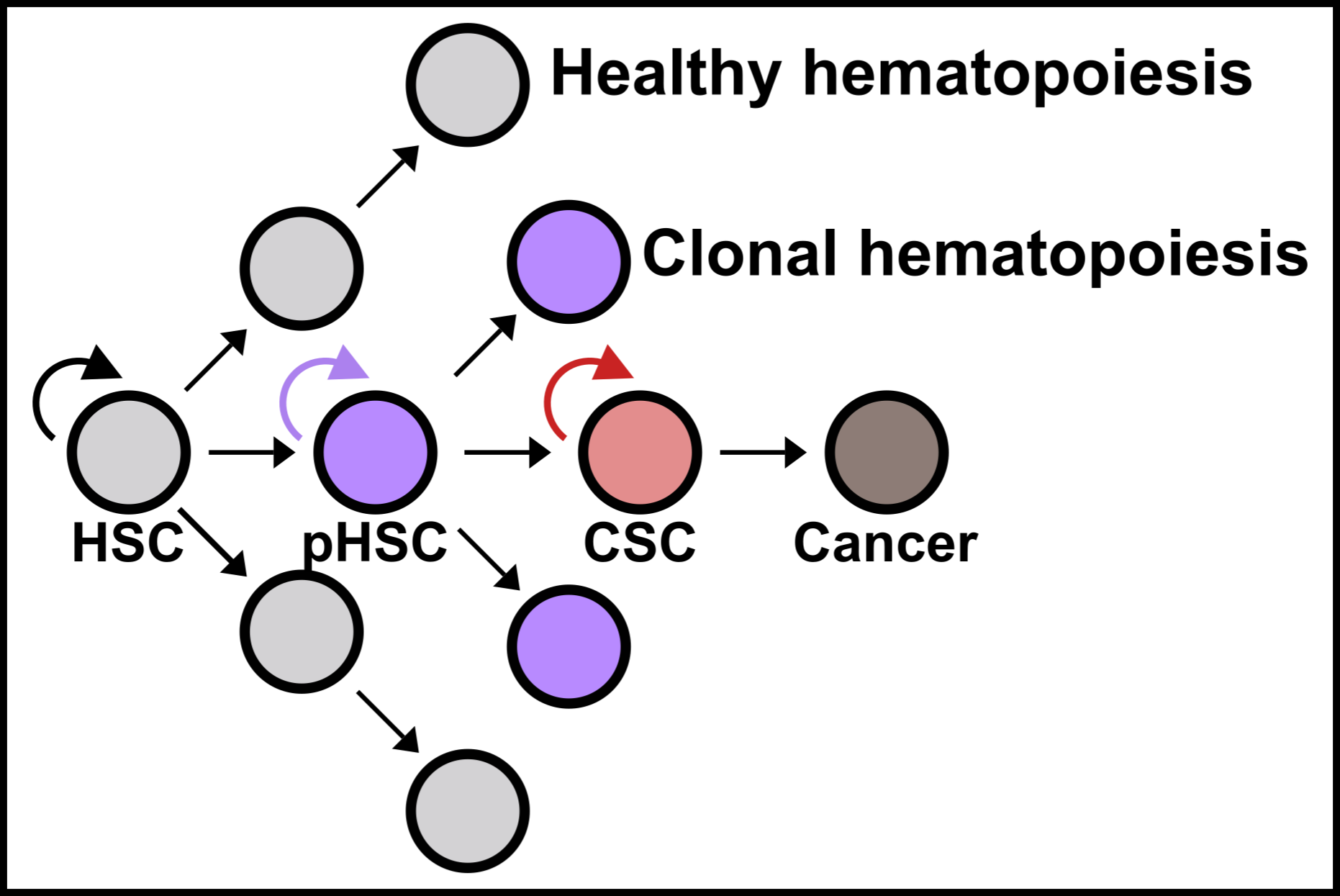
We are now using these methods to develop an integrative, multi-domain framework (Figure 3) for studying healthy and malignant hematopoiesis, with the long-term goal of designing more effective disease monitoring and treatment strategies for patients.
Specifically, we will answer several important questions using the cancer stem cell model as a reference. First, how do cancer stem cells (CSCs) emerge from healthy HSPCs, and can we identify features that distinguish CSCs from preleukemic HSCs (pHSCs) and healthy HSPCs (Project 1)? Can we use this framework to study CSCs across other myeloid neoplasms (Project 2)? Finally, can we use this information to improve clinical disease monitoring assays and develop CSC targeted therapies to cure patients (Project 3)?
Project #1: Stem cells in healthy and malignant hematopoiesis
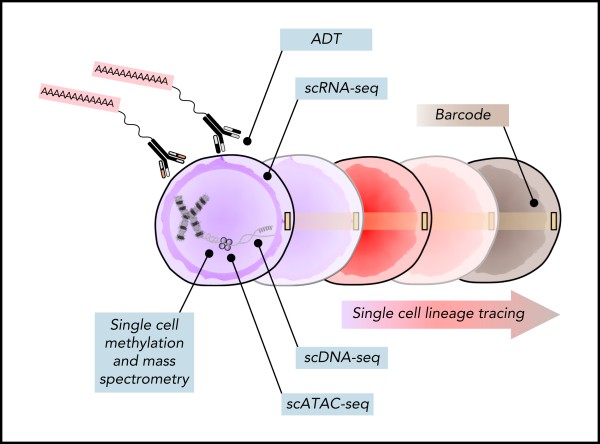
Identifying CSCs, pHSCs, and healthy HSPCs requires confirming both the clonal nature of the cell and its functional capacity in vivo. We are developing single cell methods for both lineage tracing and multi-omic profiling to enhance functional and mechanistic studies in CSC pathogenesis. Our long term goal is to use these methods to develop stem cell specific clinical biomarker assays.
Project #2: Stem cell architecture in myeloid neoplasms
Myeloid CSCs are poorly defined in myeloid cancers like myelodysplastic neoplasms (MDS), myeloproliferative neoplasms (MPN), systemic mastocytosis (SM), and chronic myelomonocytic leukemia (CMML). We are currently applying our multi-omic single cell methods to study myeloid CSCs across myeloid malignancies. Through collaborations with multiple clinical investigators at Stanford and beyond, we are acquiring pre and on-treatment samples from patients receiving standard and novel anti-cancer therapies. These samples are being profiled prospectively with the goal of defining the cellular architecture across myeloid neoplasms on treatment using both single cell and bulk molecular profiling methods and functional hematologic assays.

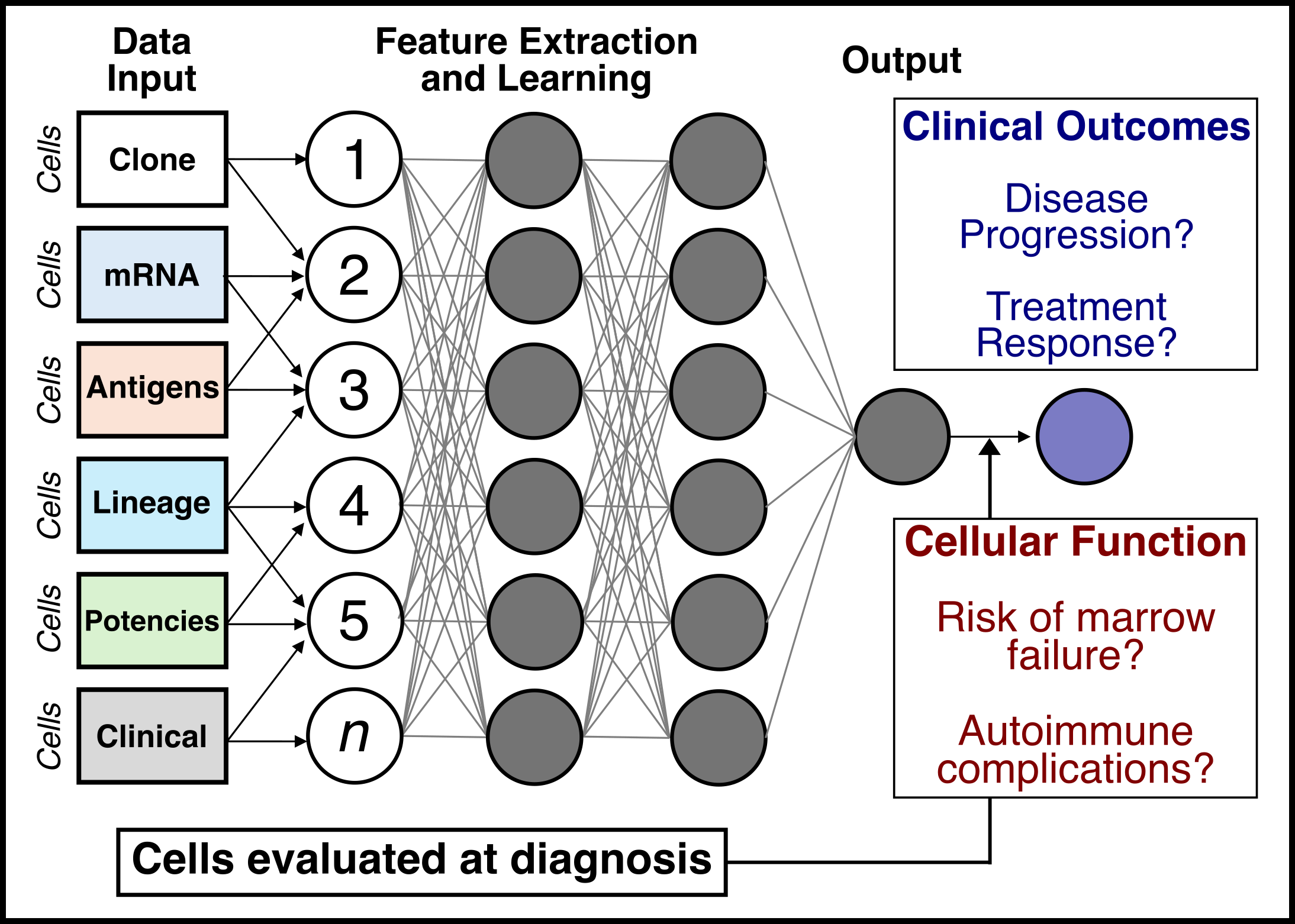
Project #3: Predictive models and clinical assays
A long-term goal is to improve the clinical management of patients with myeloid neoplasms. To address this goal, we are developing a multi-domain framework for linking neoplastic cells obtained at diagnosis and during treatment with outcomes. We are using this data to design computational models using machine learning to predict the natural history of myeloid neoplasms and its response to treatment. The goal is to develop clinical assays and predictive models to guide the study and management of myeloid cancers.